iTRAQ Quantitative Proteomics
A quantitative proteomics technique
Written by Qifeng Lin OF THE PROTEIN AND PROTEOMICS CENTRE, DEPARTMENT OF BIOLOGICAL SCIENCES, NATIONAL UNIVERSITY OF SINGAPORE
iTRAQ is a powerful quantitative proteomics technique for broad and unbiased quantification of proteins expressed in multiple biological samples. By generating relative quantitative proteome profiles, iTRAQ enables you to perform comparisons across multiple samples in a single experiment. Examples include comparisons between healthy and disease/s, drug treatments or time-course studies.
iTRAQ utilises isobaric (same mass) stable isotope reagents, each consisting of a reporter group, a balance group and an amine-reactive group. Eight different iTRAQ reagents are available, allowing for up to eight different samples to be differentially labelled and combined in a single analysis. The amine-reactive group allows the iTRAQ reagents to covalently label the primary amines (N-terminal and lysine ε-side chain) of any peptide. The eight reporter groups have masses ranging from 113 to 121 m/z, and the balance group have varying masses to maintain the same overall mass for eight reagents.
For all iTRAQ projects, please first discuss with us your experimental design and expectations.
Our typical iTRAQ 8-plex workflow can process up to eight different samples (e.g. 4 conditions, 2 replicates each). Two LCMS analysis options are available. The combined iTRAQ mixture can be directly analysed (single injection) to provide a quick and cheap overview of the dataset. Alternatively, for a deeper proteome coverage, offline fractionation (usually basic reversed phase LC) can be performed prior to the online LCMS analysis.
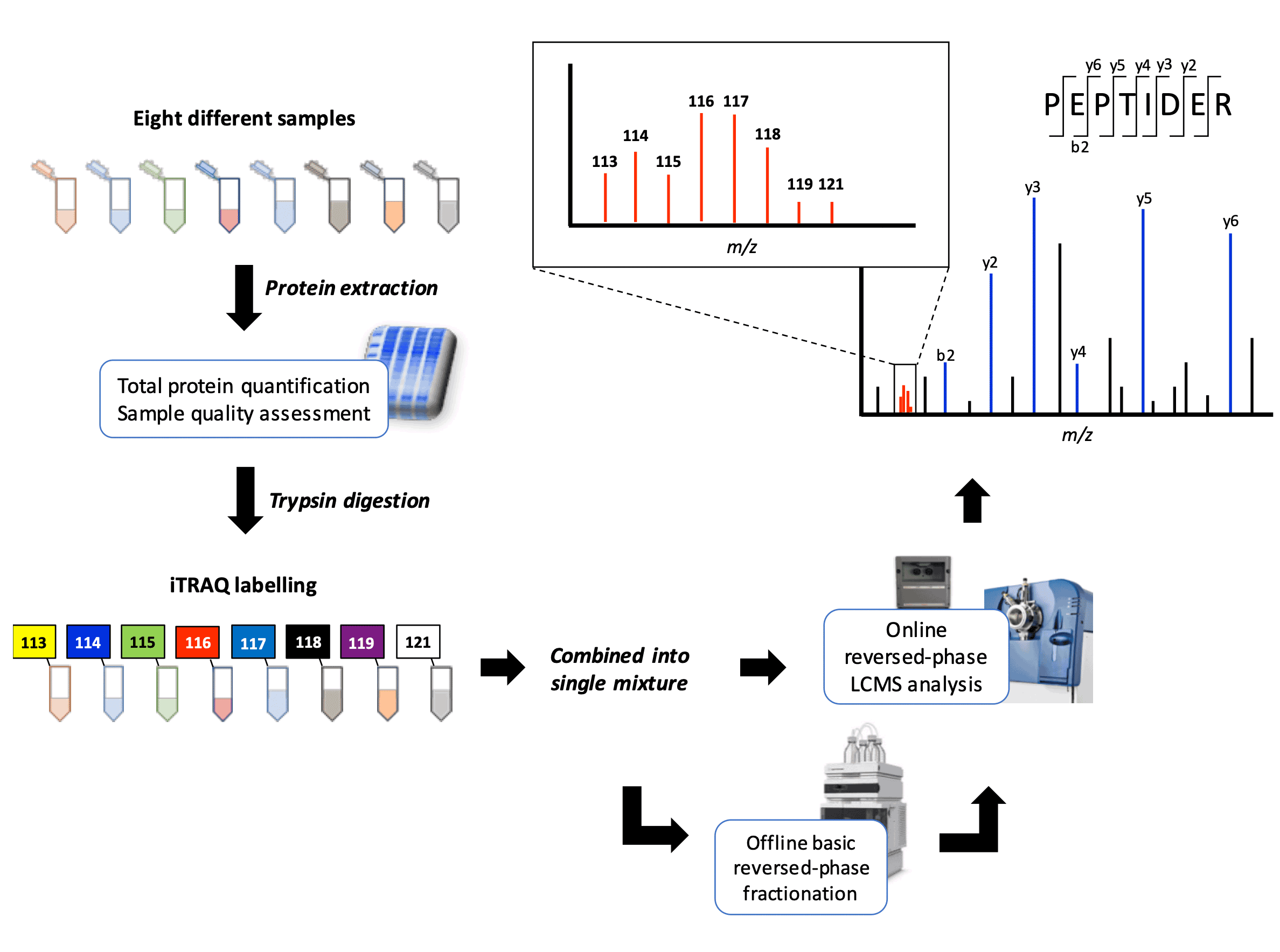
Peptide quantification is performed at the MSMS level. Labelled peptides from different samples bear the same precursor mass due to the isobaric nature of the tags. Upon fragmentation, the reporter groups are then released and generate individual peak signals. These can be analysed to infer the relative abundance of the labelled peptides in each sample. The remaining fragment ion information is then used to deduce the identity of the peptide.
Key info
- Up to eight different samples can be analysed in a single experiment
- Virtually any type of protein sample from any biological system analysable
- A reference protein database will be required
- At least 50 – 100 µg of protein for each sample recommended as starting material
- Lysis buffer should not contain primary amines (e.g. Tris), thiols or high concentrations of detergents
- Please discuss with us on the optimal lysis buffer for your biological system
- Duration depends on depth of analysis
- A single injection analysis may require 2-3 weeks
- Fractionated analyses may require up to 2 months
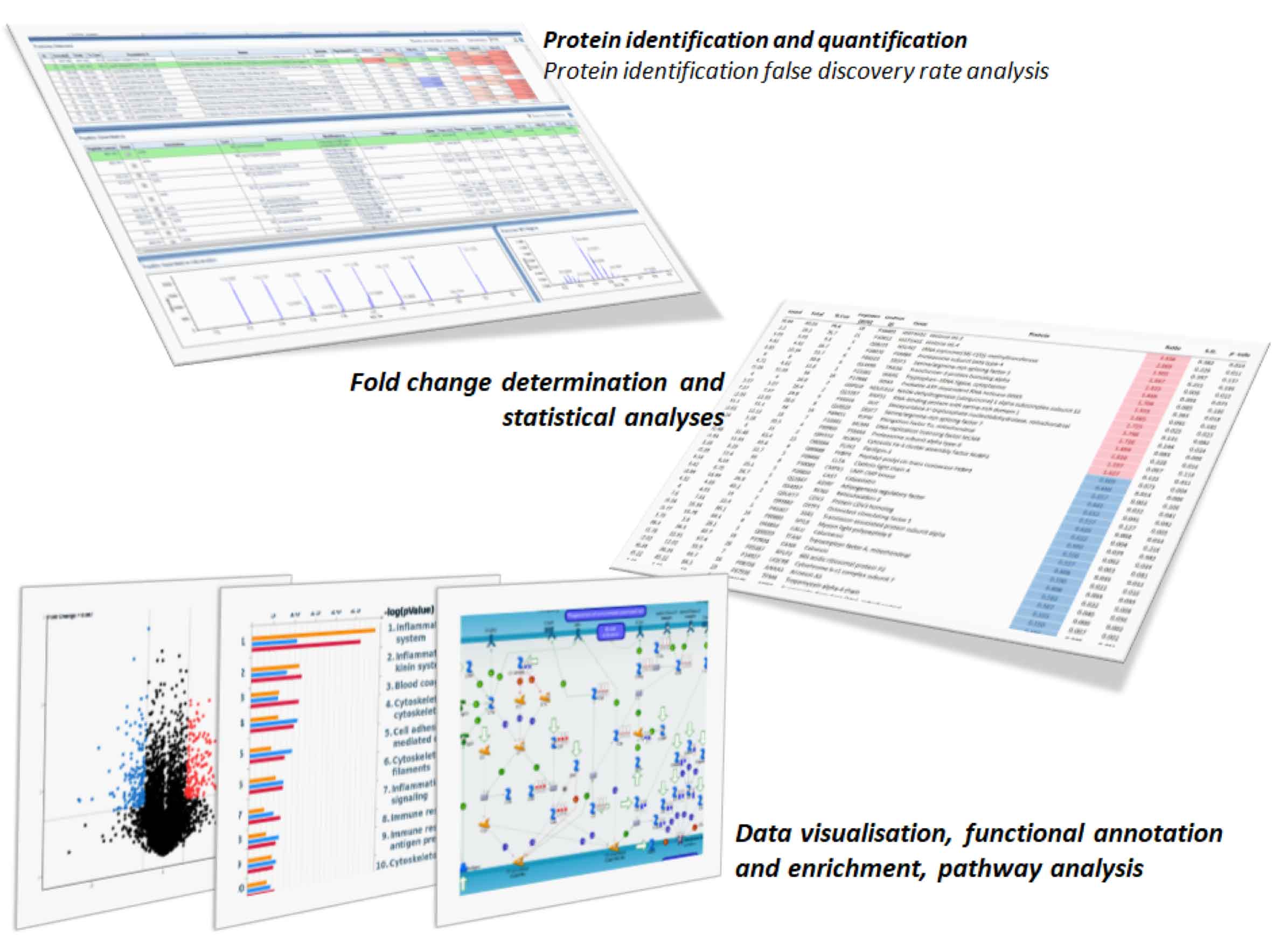
What you can expect